
It has long been recognized that the elaborate shape of neurons is fundamental to their function in the brain. We recognize that in order to move modern neuroscience toward the next critical step of integrating molecular, morphological, and connectomic properties for the unbiased classification of neuronal cell types, new genetic tools must be developed to resolve the full complexity of individual neuronal morphology. With the support of two BRAIN Initiative grants, X. William Yang's lab addressed this need for a simple, generalizable, and scalable method to sparsely label genetically-defined neuronal populations by developing reporter mouse lines conferring Cre-dependent sparse cell labeling methodology based on MOnonucleotide Repeat Frameshift (MORF) as a stochastic translational switch. The suite of MORF reporter mice labels about 1-5% of Cre+ neurons and glia distributed stochastically throughout the brain and can be imaged with endogenous fluorescence (mNeonGreen in MORF1 and EGFP in TIGRE-MORF) or stained for a multivalent immunoreporter (Spaghetti Monster fluorescent protein V5, or smFP-V5, in MORF3). With MORF, we now have the technology to label and reconstruct thousands genetically defined cells per brain for large-scale, unbiased classification and quantitative analyses of CNS cell types brainwide.
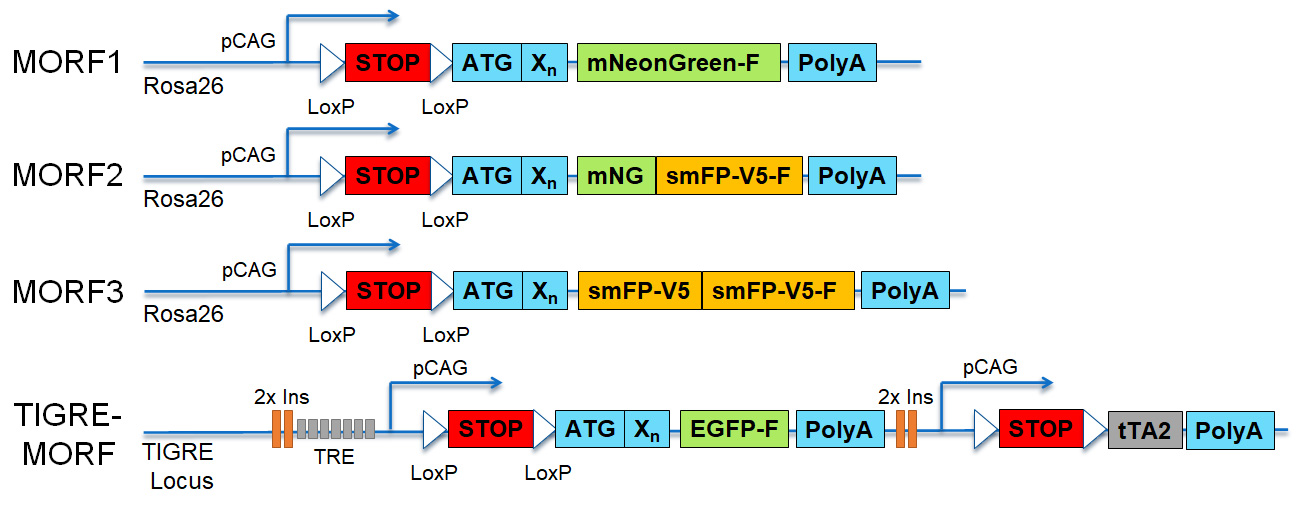
MORF knockin transgenic mouse lines include the strong, ubiquitous CAG promoter, LoxP-STOP-LoxP cassette for Cre-dependent expression, the MORF sequence between the start codon (ATG) and the reporter coding sequence, and membrane-associated reporters for brightly labeling neuron morphology with endogenous fluorescence (mNeonGreen-F, mNG-F; or EGFP-F) or following immunostaining (spaghetti monster Fluorescent Protein V5, smFP-V5-F). The MORF method utilizes an unstable, out-of-frame mononucleotide repeat that can undergo frameshift that sparsely and stochastically switch on reporter gene expression. MORF1-3 are ROSA26 locus knockins while the TIGRE-MORF is knocked into the TIGRE locus and harbors a tTA2-TRE transcription mediated amplification cassette for maximum reporter expression in additions to insulator sequences (2x Ins).
The endogenous florescence of MORF1 and TIGRE-MORF can be imaged directly. The smFP-V5 epitope tag of the MORF3 can be visualized using conventional immunostaining protocols with thin sections or can be cleared and immunostained with thick tissue sections and whole brain using iDISCO+. Summarized and detailed protocol listed below:
The following MORF reporter lines have been assigned stock numbers and are currently in the process of being deposited in the Jackson Laboratory (JAX):
These MORF lines are now listed on the JAX website (https://www.jax.org/search?q=